Assessment |
Biopsychology |
Comparative |
Cognitive |
Developmental |
Language |
Individual differences |
Personality |
Philosophy |
Social |
Methods |
Statistics |
Clinical |
Educational |
Industrial |
Professional items |
World psychology |
Biological: Behavioural genetics · Evolutionary psychology · Neuroanatomy · Neurochemistry · Neuroendocrinology · Neuroscience · Psychoneuroimmunology · Physiological Psychology · Psychopharmacology (Index, Outline)
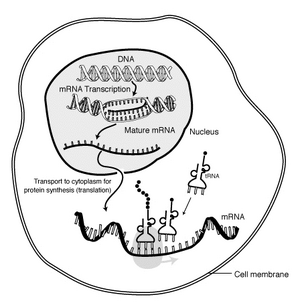
The "life cycle" of an mRNA in a eukaryotic cell. RNA is transcribed in the nucleus; once completely processed, it is transported to the cytoplasm and translated by the ribosome. At the end of its life, the mRNA is degraded.
Messenger Ribonucleic Acid (mRNA) is RNA that encodes and carries information from DNA during transcription to sites of protein synthesis to undergo translation in order to yield a gene product.
mRNA "life cycle"[]
The brief life of an mRNA molecule begins with transcription and ultimately ends in degradation. During its life, an mRNA molecule may also be processed, edited, and transported prior to translation. Eukaryotic mRNA molecules often require extensive processing and transport, while prokaryotic molecules do not.
Transcription[]
- Main article: Transcription (genetics)
During transcription, RNA polymerase makes a copy of a gene from the DNA to mRNA as needed. This process is similar in eukaryotes and prokaryotes. One notable difference, however, is that eukaryotic RNA polymerase associates with mRNA processing enzymes during transcription so that processing can proceed quickly after the start of transcription. The short-lived, unprocessed or partially processed, product is termed pre-mRNA; once completely processed, it is termed mature mRNA.
Eukaryotic pre-mRNA processing[]
- Main article: Post transcriptional modification
Processing of mRNA differs greatly between eukaryotes and prokaryotes. Prokaryotic mRNA is essentially mature upon transcription and requires no processing, except in rare cases. Eukaryotic pre-mRNA, however, requires extensive processing.
Splicing[]
- Main article: Splicing (genetics)
Splicing is the process by which pre-mRNA is modified to remove certain stretches of non-coding sequences called introns; the stretches that remain include protein-coding sequences and are called exons. Sometimes pre-mRNA messages may be spliced in several different ways, allowing a single gene to encode multiple proteins. This process is called alternative splicing. Splicing is usually performed by an RNA-protein complex called the spliceosome, but some RNA molecules are also capable of catalyzing their own splicing (see ribozymes).
5' cap addition[]
- Main article: 5' cap
The 5' cap is modified guanine nucleotide is added to the "front" (5' end) of the pre-mRNA. This modification is critical for recognition and proper attachment of mRNA to the ribosome, as well as protection from 5' exonucleases. It may also be important for other essential processes, such as splicing and transport.
Polyadenylation[]
- Main article: Polyadenylation
Polyadenylation is the covalent linkage of a polyadenylyl moiety to a messenger RNA molecule. In eukaryotic organisms, polyadenylation is the mechanism by which most messenger RNA (mRNA) molecules are terminated at their 3' ends. The poly(A) tail aids in mRNA stability by protecting it from exonucleases. Polyadenylation is also important for transcription termination, export of the mRNA from the nucleus, and translation. Some prokaryotic mRNAs also are polyadenylated, although the poly(A) tail's function is different from that in eukaryotes.
Polyadenylation occurs during and immediately after transcription of DNA into RNA. After transcription has been terminated, the mRNA chain is cleaved through the action of an endonuclease complex associated with RNA polymerase. The cleavage site is characterized by the presence of the base sequence AAUAAA near the cleavage site. After the mRNA has been cleaved, 80 to 250 adenosine residues are added to the free 3' end at the cleavage site. This reaction is catalyzed by polyadenylate polymerase.
Editing[]
In some instances, an mRNA will be edited, changing the nucleotide composition of that mRNA. An example in humans is the apolipoprotein B mRNA, which is edited in some tissues, but not others. The editing creates an early stop codon, which upon translation, produces a shorter protein.
Transport[]
Another difference between eukaryotes and prokaryotes is mRNA transport. Because eukaryotic transcription and translation is compartmentally separated, eukaryotic mRNAs must be exported from the nucleus to the cytoplasm. Mature mRNAs are recognized by their processed modifications and then exported through the nuclear pore.
Translation[]
- Main article: Translation (genetics)
Because prokaryotic mRNA does not need to be processed or transported, translation by the ribosome can begin immediately after the start of transcription. Therefore, it can be said that prokaryotic translation is coupled to transcription and occurs co-transcriptionally.
Eukaryotic mRNA that has been processed and transported to the cytoplasm (i.e. mature mRNA) can then be translated by the ribosome. Translation may occur at ribosomes free-floating in the cytoplasm, or directed to the endoplasmic reticulum by the signal recognition particle. Therefore, unlike prokaryotes, eukaryotic translation is not directly coupled to transcription.
Degradation[]
After a certain amount of time the message degrades into its component nucleotides, usually with the assistance of RNases. Due to mRNA processing, eukaryotic mRNAs are generally more stable than prokaryotic mRNAs.
mRNA structure[]

The structure of a mature eukaryotic mRNA. A fully processed mRNA includes a 5' cap, 5' UTR, coding region, 3' UTR, and poly(A) tail.
5' cap[]
- Main article: 5' cap
A 5' cap, also termed an RNA cap, an RNA 7-methylguanosine cap or an RNA m7G cap, is a modified guanine nucleotide that has been added to the "front" or 5' end of the messenger RNA shortly after the start of transcription. The 5' cap consists of a terminal 7-methylguanosine residue which is linked through a 5'-5'-triphosphate bond to the first transcribed nucleotide. Its presence is critical for recognition by the ribosome and protection from RNases.
Cap addition is coupled to transcription, and occurs co-transcriptionally, such that each influences the other. Shortly after the start of transcription, the 5' end of the mRNA being synthesized is bound by a cap-synthesizing complex associated with RNA polymerase. This enzymatic complex catalyzes the chemical reactions that are required for mRNA capping. Synthesis proceeds as a multi-step biochemical reaction.
First, the triphosphate at the 5' end of the newly synthesized RNA is cleaved. The enzyme phosphohydrolase cleaves the gamma phosphodiester bonds while leaving the alpha and beta phosphates. Second, the enzyme guanylyltransferase transfers a guanine and its alpha phosphate onto the beta phosphate of the 5' end of the mRNA producing a 5'-5'-triphosphate linkage. Third, the nitrogen-7 (N-7) position of the newly added guanine is methylated (guaninemethylation) by the enzyme guanine-7-methyltransferase. Finally, 2'-O-methyltransferase methylates the 2' position of the ribose sugar. This methyl group provides extra stability to the RNA due to the protection from phosphoester cleavage by nucleophilic attack of the neighbor hydrogen. After the 5' end has been capped, it is released from the cap-synthesizing complex and is subsequently bound by a cap-binding complex associated with RNA polymerase.
Coding regions[]
Coding regions are composed of codons, which are decoded and translated into protein by the ribosome. Coding regions begin with the start codon and end with the one of three possible stop codons. In addition to protein-coding, portions of coding regions may also serve as regulatory sequences as exonic splicing enhancers or exonic splicing silencers.
Monocistronic versus Polycistronic mRNA[]
A mRNA molecule is said to be monocistronic when it contains the genetic information to translate only a single protein. This is the case for most of the eukaryotic mRNAs[1].
On the other hand, polycistronic mRNA carries the information of several proteins, which are translated into single proteins. Most of the mRNA found in prokaryotes is polycistronic[1]. An example of polycistronic mRNA is in the biosynthetic pathway for tryptophan in E.Coli: a single mRNA molecule about 7000 nucleotides long specifies five enzymes, each of which has its own start and stop signals on the mRNA. Another classic example is found in the lac operon, where the lacZ, lacY and lacA genes are transcribed into a single polycistronic mRNA.
Monocistronic mRNA are in some cases (e.g. viruses) translated to form a polyprotein, which is processed by proteolytic cleavage to form the mature gene products.
Examples of polycistronic mRNA in eukaryotes are often found in the chloroplasts of vascular plants[2]. This notable exception to the otherwise monocistronic nature of nuclear genes in eukaryotes may be interpreted as evidence suporting the endosymbiotic theory.
Untranslated regions[]
- Main article: 5' UTR
Untranslated regions (UTRs) are sections of the RNA before the start codon and after the stop codon that are not translated, termed the five prime untranslated region (5' UTR) and three prime untranslated region (3' UTR), respectively. These regions are transcribed as part of the same transcript as the coding region. Several roles in gene expression have been attributed to the untranslated regions, including mRNA stability, mRNA localization, and translational efficiency. The ability of a UTR to perform these functions depends on the sequence of the UTR and can differ between mRNAs.
Stability of mRNAs may be mediated by the 5' UTR and 3' UTR due to varying affinity for certain RNA degrading enzymes called ribonucleases, which can promote or inhibit the relative stability of the RNA molecule. The greater the stability of an mRNA, the more protein that may be produced from that transcript.
Cytoplasmic localization of mRNA is thought to be a function of the 3' UTR. Proteins that are needed in a particular region of the cell can actually be translated there; in such a case, the 3' UTR may contain sequences that allow the transcript to be localized to this region for translation.
Translational efficiency, and even inhibition of translation altogether, can be mediated by UTRs. Proteins that bind to either the 3' or 5' UTR may affect translation by interfering with the ribosome's ability to bind to the mRNA.
Some of the elements contained in untranslated regions form a characteristic secondary structure when transcribed into RNA. These structural mRNA elements are involved in regulating the mRNA. Some, such as the SECIS element, are targets for proteins to bind. One class of mRNA element, the riboswitches, directly bind small molecules, changing their fold to modify levels of transcription or translation. In these cases, the mRNA regulates itself.
3' poly(A) tail[]
- Main article: Polyadenylation
The 3' poly(A) tail is a long sequence of adenine nucleotides (often several hundred) added to the "tail" or 3' end of the pre-mRNA through the action of an enzyme, polyadenylate polymerase. The poly(A) tail is added on to the transcripts that contain a specific sequence, the AAUAAA signal. The importance of the AAUAAA signal is demonstrated by a mutation in the human alpha 2-globin gene which mutates the original sequence AATAAA into AATAAG, which can lead to hemoglobin deficiencies.[3]
Anti-sense mRNA[]
During transcription, double stranded DNA produces mRNA from the sense strand; the other, complementary, strand of DNA is termed anti-sense. Anti-sense mRNA is an RNA complementary in sequence to one or more mRNAs. In some organisms, the presence of an anti-sense mRNA can inhibit gene expression by base-pairing with the specific mRNAs. In biochemical research, this effect has been used to study gene function, by simply shutting down the studied gene by adding its anti-sense mRNA transcript. Such studies have been done on the worm Caenorhabditis elegans and the bacterium Escherichia coli. This plays a part in RNA interference.
See also[]
References[]
- ↑ 1.0 1.1 Kozak, M. (March 1983). Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles.. Microbiological Reviews 47 (1): 1-45. PMID 6343825.
- ↑ Drapier D, Suzuki H, Levy H, Rimbault B, Kindle KL, Stern DB, Wollman FA (June 1998). The chloroplast atpA gene cluster in Chlamydomonas reinhardtii. Functional analysis of a polycistronic transcription unit.. Plant physiology 117 (2): 629-641. PMID 9625716.
- ↑ Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ. (1983). α-thalassaemia caused by a polyadenylation signal mutation. Nature 306 (5941): 398–400. PMID 6646217 DOI:10.1038/306398a0.
Links[]
Life of mRNA Flash animation
| Nucleic acids edit |
|---|
| Nucleobases: Adenine - Thymine - Uracil - Guanine - Cytosine - Purine - Pyrimidine |
| Nucleosides: Adenosine - Uridine - Guanosine - Cytidine - Deoxyadenosine - Thymidine - Deoxyguanosine - Deoxycytidine |
| Nucleotides: AMP - UMP - GMP - CMP - ADP - UDP - GDP - CDP - ATP - UTP - GTP - CTP - cAMP - cGMP |
| Deoxynucleotides: dAMP - dTMP - dUMP - dGMP - dCMP - dADP - dTDP - dUDP - dGDP - dCDP - dATP - dTTP - dUTP - dGTP - dCTP |
| Nucleic acids: DNA - RNA - LNA - PNA - mRNA - ncRNA - miRNA - rRNA - siRNA - tRNA - cDNA - snRNA - snoRNA - mtDNA - Oligonucleotide
|
da:MRNA de:MRNA es:ARN mensajero fr:Acide ribonucléique messager he:MRNA la:MRNA lt:IRNR nl:Messenger RNA pt:ARN mensageiro sv:Budbärar-RNA vi:RNA thông tin zh:MRNA Template:Messenger RNA